GWAS summary data ecosystem
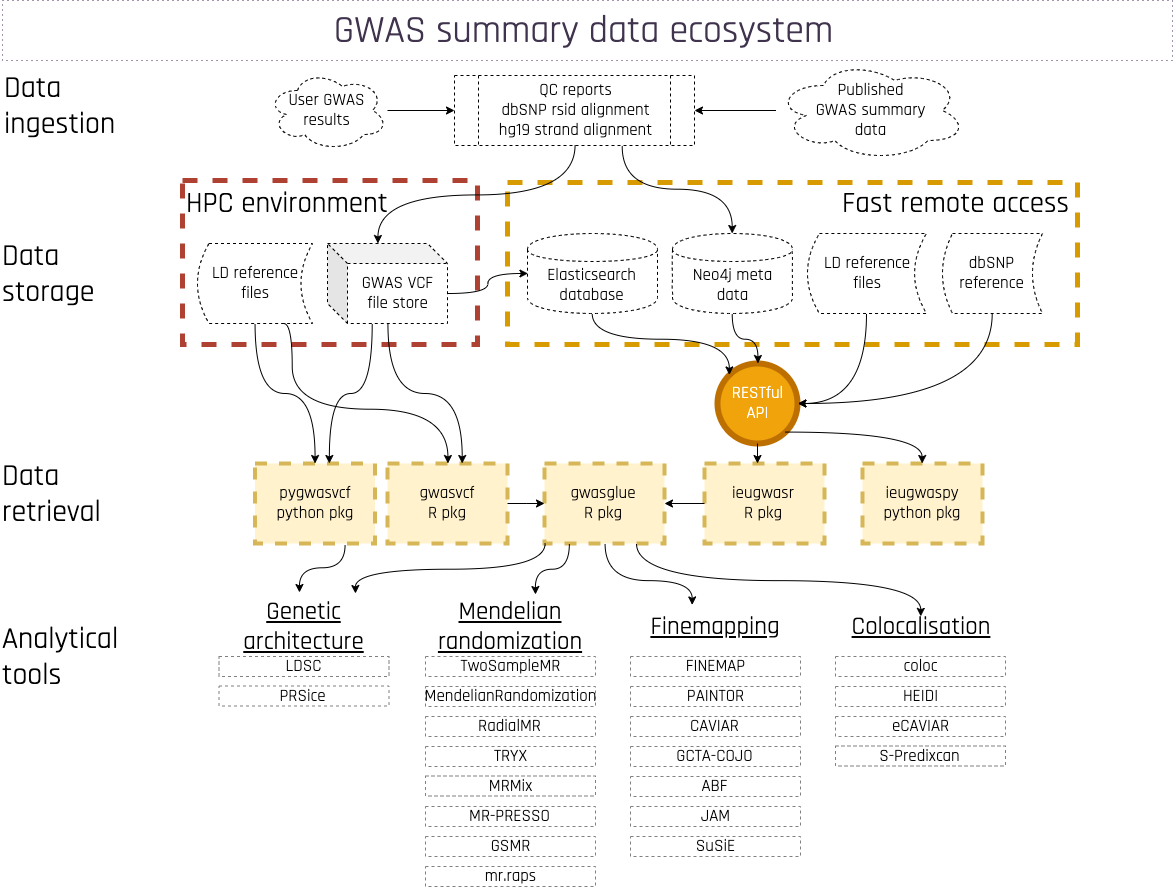
Background
Developed at the MRC Integrative Epidemiology Unit (IEU) at the University of Bristol, this resource is a manually curated collection of complete GWAS summary datasets made available as open source files for download, or by querying a database of the complete data.
This project began as the underlying database for the MR-Base and LD Hub projects. These data now serve as an input source to a wider number of analytical tools that implement methods such as Mendelian randomization, fine mapping, colocalisation, GWAS visualisation etc. Please see the API page for a list of R and python packages that will connect to the data.
The database comprises mainly publicly available datasets, but also includes a number of private datesets whose access is controlled through OAuth2.0 authentication. Please contact us if you have datasets that you would like to add, whether public or private.
The GWAS VCF format
We have made all the public data available for download. We are using the GWAS VCF format to store the GWAS summary data to ensure alignment with the hg19 reference sequence, and to enable very fast querying. More information is available here: https://github.com/MRCIEU/gwas-vcf-specification and on biorxiv.
Citing this resource
Please ensure that the original data are cited whenever it is used. Pubmed IDs are made available through the meta data.
-
If you access the data through the OpenGWAS database API:
The MRC IEU OpenGWAS data infrastructure. Ben Elsworth, Matthew Lyon, Tessa Alexander, Yi Liu, Peter Matthews, Jon Hallett, Phil Bates, Tom Palmer, Valeriia Haberland, George Davey Smith, Jie Zheng, Philip Haycock, Tom R Gaunt, Gibran Hemani. bioRxiv 2020.08.10.244293v1. doi: 10.1101/2020.08.10.244293
The MR-Base platform supports systematic causal inference across the human phenome. Hemani G, Zheng J, Elsworth B, Wade KH, Baird D, Haberland V, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC, The MR-Base Collaboration. eLife 2018;7:e34408. doi: 10.7554/eLife.34408 If using GWAS-VCF files:
The variant call format provides efficient and robust storage of GWAS summary statistics. Matthew Lyon, Shea J Andrews, Ben Elsworth, Tom R Gaunt, Gibran Hemani, Edoardo Marcora. bioRxiv 2020.05.29.115824; doi: https://doi.org/10.1101/2020.05.29.115824
Bulk downloads
OpenGWAS on Oracle Open Data
OpenGWAS VCF files are now also hosted on Oracle Open Data as part their initiative to openly and freely share datasets for research, education, and development. Sample scripts are provided on their site to demonstrate how to download data.
Direct download from OpenGWAS servers
You can also download VCF files directly from our servers. There is not currently a single file to download everything - it would be extremely large (many terrabytes) and constantly out of date as we continually add new studies. To programmatically download a file you can use the wget command. For example, if you know the ID of the GWAS dataset (ukb-b-19953 in the example below) then the following commands will download that file and its index:
wget https://gwas.mrcieu.ac.uk/files/ukb-b-19953/ukb-b-19953.vcf.gz
wget https://gwas.mrcieu.ac.uk/files/ukb-b-19953/ukb-b-19953.vcf.gz.tbi
A list of all datasets, along with their meta-data and IDs, is available through the API, R or python packages, so it should be straightforward to create a subset of datasets to be downloaded through a loop. If doing so, please be considerate and avoid doing this type of activity in parallel as it could impact our servers and make the resource difficult to access for others.
To learn more about how to use GWAS-VCF files, including how to use R and python to efficiently manipulate and query, or convert to text files, see here.
Credits
Many people have contributed to the IEU GWAS database project. Main parties are:
- Ben Elsworth
- Chris Zheng
- Gibran Hemani
- Jon Hallett
- Matt Lyon
- Peter Matthews
- Philip Haycock
- Tessa Alexander
- Tom Gaunt
- Valeriia Haberland
- Yi Liu
Sincere thanks also go to the many GWAS consortia who have made the GWAS data that they generated publicly available, and many members of the IEU who have contributed to curating these data.
Dependencies
The IEU GWAS database project is built upon many important open source software projects.
Contributing consortia
Many thanks to the NHGRI-EBI GWAS catalog and the studies contributing data to OpenGWAS
- ADIPOGen
- IIBDGC
- GIANT
- SSGAC
- ReproGen
- MESA
- HaemGen
- CORNET
- GliomaScan
- DCCT / EDIC
- BioBank Japan
- IMSGC
- IAC
- EAGLE
- EGG
- ENIGMA
- GIS
- GUGC
- HRgene
- MinE
- UK Biobank
- CKDGen
- ISGC
- IPSCSG
- OCAC
- BCAC
- GPC
- CHARGE
- arcOGEN
- PRACTICAL
- PGC
- iPSYCH-PGC
- PGC-ED
- GeM-HD
- Sclerostin genetics consortium
- UK Biobank+INTERVAL+UK BiLEVE
- Biobank Japan Project
- DIAGRAM
- DIAGRAMplusMetabochip
- IGAP
- GLGC
- AMDGene
- ALSgene
- GABRIEL
- GCAN
- GEFOS
- C4D
- CARDIoGRAMplusC4D
- MAGIC
- CARDIoGRAM
- PanScan
- ICBP
- TAG
- ILCCO
- SpiroMetaplusCHARGE
- TRICL